4th April 25
Image classification by evolving bytecode
We investigate the potential of evolving the bytecode of a biologically-inspired virtual machine as a plausible strategy for machine learning. We simulate evolution with the Zyme language and strand-based virtual machine. Our test problem is classifying handwritten digits from a subset of the MNIST dataset. Beginning with an initial program with performance no better than random guessing, we achieve consistent accuracy improvements through random mutations over 50 generations. Although these results fall short of state-of-the-art methods like neural networks, they demonstrate that adaptive mutations are found consistently and suggest the potential for evolving Zyme bytecode to competitively tackle the full MNIST task. This result also suggests the value of alternative virtual machines architectures in genetic programming, particularly those optimized for evolvability.
1. Introduction
Evolution's creative potential is evident not only in the remarkable biodiversity across Earth, but also in the unique adaptations species find to common environmental challenges. This ability to generate innovative solutions has captivated researchers since the early days of computing1, inspiring the idea that an artificial evolutionary process could generate programs for specific tasks without being explicitly coded to do so.
The evolution of programs is far more than a whimsical idea though - if realized, it's potentially an approach to machine learning and artificial intelligence. John Koza, founder of the program-evolving discipline known as genetic programming, captured this sentiment succinctly, noting: “virtually any problem in artificial intelligence, symbolic processing, and machine learning can be viewed as requiring discovery of a computer program that produces some desired output for particular inputs”2.
Genetic programming uses a combination of supervised learning techniques and trial and error to mimic natural selection. Programs undergo rounds of random modifications (mutations) and are then evaluated to determine the degree to which they produce the desired output. The top-performing programs are selected for further rounds of mutation and evaluation. The hope is that after multiple generations, this process will produce a program that generates the expected output from a set of unseen inputs with high fidelity.
Despite its conceptual appeal, genetic programming falls far short of modern machine learning methods, particularly neural networks. This disparity is illustrated by the MNIST handwritten digit classification task, which involves identifying the digit (0–9) in 28x28 pixel grayscale images. MNIST is sufficiently straightforward that it serves as an introductory exercise for neural networks, consistently achieving over 99% accuracy, yet remains virtually unsolvable through genetic programming3.
Traditional approaches rely on program representations derived from C, Java, Python, and other languages used in real-world software development. These languages as well as the computational architectures or virtual machines they run on are optimized for programmability, efficiency, and reliability. Built with a human-centric philosophy, the priority is systems that are predictable and logical, enabling developers to infer program behavior directly from source code while minimizing the risk small mistakes could cause unpredictable and undetectable changes in program behaviour. This rigidity is antithetical to principles of evolution, rendering them fragile to random mutations. Sadly, these human centered design decisions actively undermine the flexibility and adaptability essential for evolutionary processes.
To overcome the limitations of human-oriented languages and competitively solve MNIST, we developed Zyme, an evolution-oriented programming language and virtual machine specifically designed for genetic programming4. Zyme is inspired by the molecular processes in living cells. It consists of two components: a language/compiler that generates a bytecode format engineered to be evolvable, and a unique virtual machine built to mimic an abstract cellular metabolism.
Here, we demonstrate the potential of Zyme to solve a small subset of the MNIST dataset, containing only four digits (0, 1, 2, and 3). Beginning with an initial program whose performance is no better than random guessing, we achieve consistent accuracy improvement, accumulating an increase of up to ~40% over 50 generations. Notably, Zyme’s instruction set is restricted to only byte-level operations with no built-in image-processing functions5, meaning all progress stems from the system’s ability to autonomously derive meaningful features directly from raw binary data. These results suggest that evolution-oriented languages like Zyme could be generalisable across tasks, and if scalable, serve as a foundation for machine learning through genetic programming.
2. Results
To illustrate machine learning through the evolution of Zyme programs, we applied the approach to the MNIST dataset for image classification. We will evolve a Zyme program which takes pixel image data (raw binary arrays) as input and returns a predicted label (a single byte) as output (see Methods 3.1 for details of Zyme bytecode execution). The program's accuracy will be evaluated by testing it on a set of input-output pairs - images with their corresponding labels - and measuring how often it correctly predicts the label.
The current implementation of the Zyme virtual machine is fully functional but remains in a prototype state, making it relatively slow, and limits the scale of preliminary experiments. Thus, we focused on a tiny subset of the MNIST dataset, consisting of four digits (0, 1, 2, and 3), to quickly determine whether there is any potential for learning rather than solving MNIST in its entirety (see Methods 3.2).
Zyme bytecode is evolved as a population of programs, with the starting population consisting of 50 identical copies of a hand-crafted initial program (see Methods 3.3). The evolutionary process follows a two step approach: (1) reproduction with random mutation then (2) selection, where the top performing programs are selected to make up the population of the next generation (see Methods 3.4).
We perform four replicates of this evolutionary process, each running for 50 generations and starting from the same initial population of programs, to assess robustness and reproducibility. Upon completion, we assess the accuracy of each program within every population at each generation using the test dataset (rather than batches of training dataset) to measure their ability to generalize to unseen examples (shown in Figure 1).
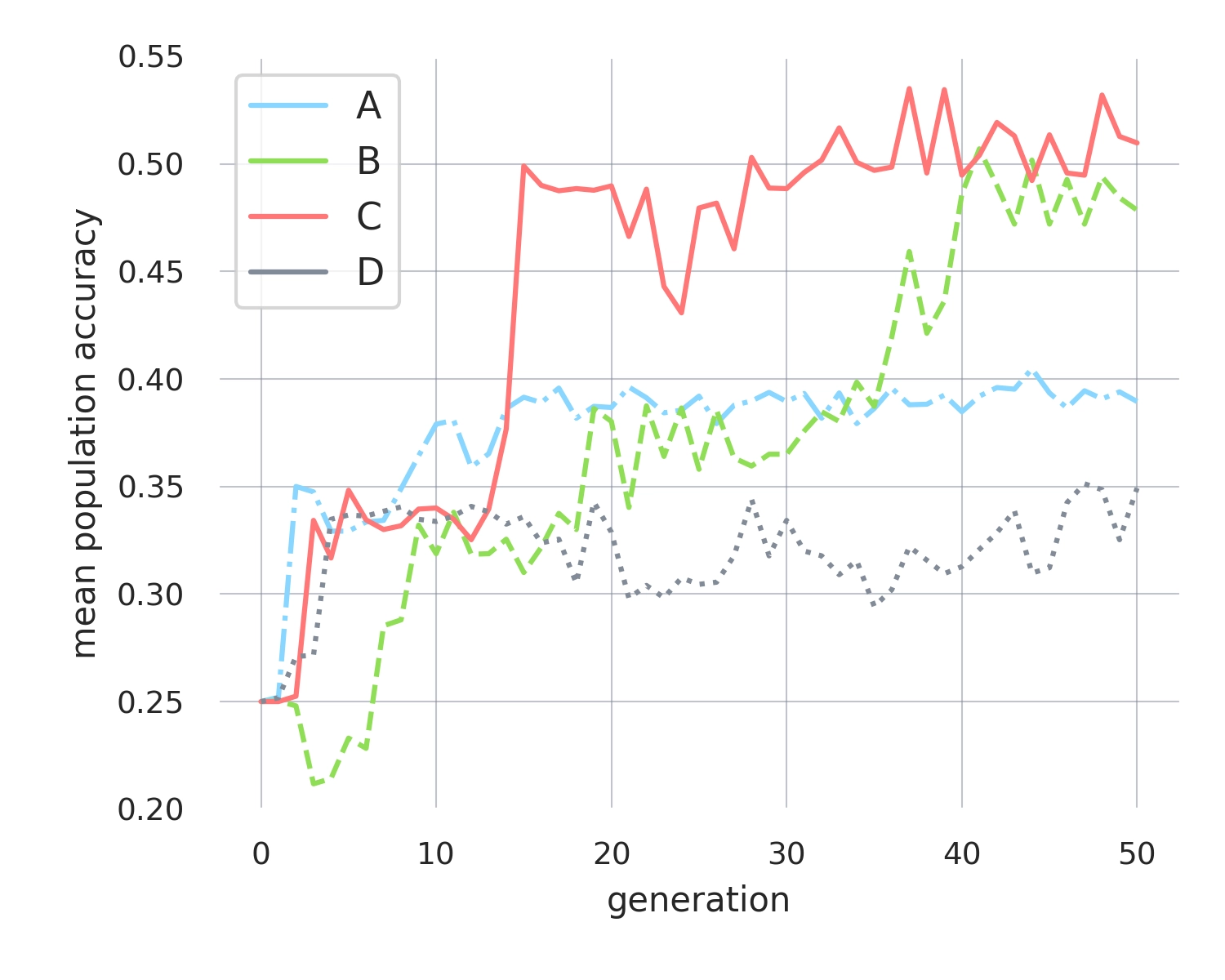
Across the replicates, we observe a consistent increase in accuracy, with the final populations of programs achieving accuracy up to ~50%, representing an improvement of 25% compared to the initial random-level performance programs. Although these results do not equate to ‘solving’ MNIST, they clearly demonstrate that individual mutations in Zyme bytecode can be sufficiently adaptive to yield to measurable performance gain. However, it remains unclear whether these adaptive mutations reflect the exploration of distinct algorithms or are merely trivial adjustments to the initial handcrafted program, leaving limited potential for further performance improvements.
Two of the replicate populations achieve only 35-40% accuracy, highlighting the risk of populations getting stuck and stagnating. Yet, the fact that these populations stabilize at different performance levels suggests that each is uncovering separate mechanisms to enhance performance. Further, the adaptations observed in the more successful populations—those reaching over 50% accuracy—appear to achieve this through incremental gains, where adaptations build on one another. For example, replicate B advance in three steps from 0.25 to 0.33 to 0.37 and up to 0.49. Again the intermediary steps stabilise at unique accuracy levels. This behavior is inconsistent with mutations that merely tweak a few constants in the initial handcrafted program, as such changes would likely cause all populations to converge to similar performance levels.
A closer examination of individual populations (as shown in Figure 2) reveals that some individuals achieve even higher accuracy. For instance, in replicate C, some achieve 65% accuracy, representing a 40% increase over baseline and a 15% improvement compared to the population mean. This indicates that further progress is possible, and the current top population accuracies do not represent the upper limit. However, it remains unclear why the population does not immediately jump to this higher level. We hypothesize that this is due to a mismatch between a program's performance in a specific batch (which determines selection) and its overall performance on the test examples (as shown in the figures). We observe numerous programs (though data is not shown here) that exhibit high test accuracy but relatively low batch accuracy. In such cases, selection does not favor these programs, causing the overall population performance to stall. While this may seem inefficient, we believe it helps prevent overfitting.
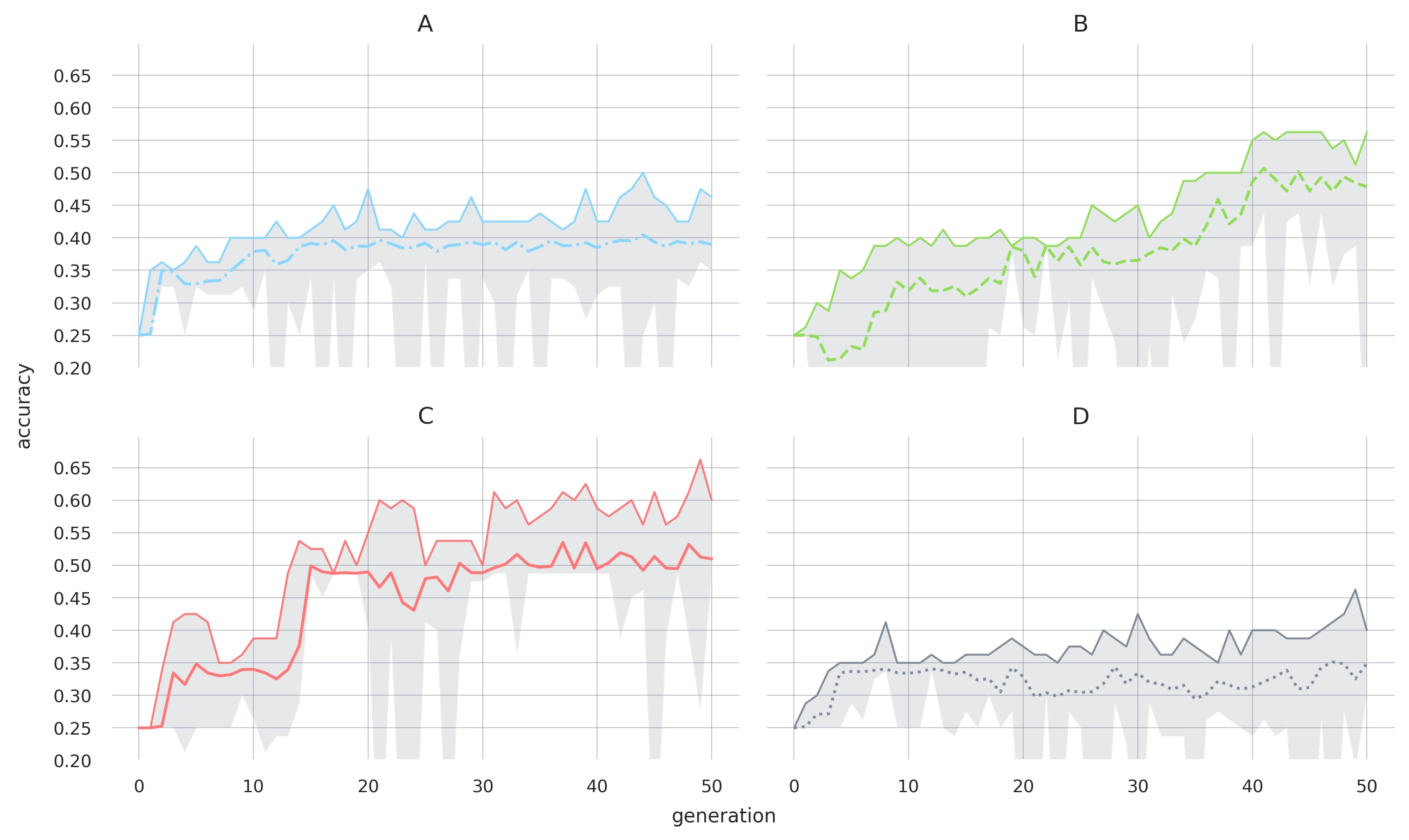
3. Methods
3.1 Running a Zyme program
Zyme programs execute on a specialized virtual machine that implements a strand-based programming paradigm6 - an architecture designed for evolvability where execution never fails and always advances. The primitive data structure is a strand, which is just an array of bytes. These strands are interpreted by the virtual machine in two ways: as code, representing a sequence of instructions known as operator strands, or as data, representing a sequence of integer values called data strands. The virtual machine’s state transitions are dictated by a set of well-defined rules that governs how the strands interact with one another.
A program is the initial state of the virtual machine, which is defined by a set of operator strands. The program bytecode is simply all of these operator strands (which again, are just arrays of bytes) combined into a single binary block with empty bytes (0x00) serving as separators. Thus, when a program is randomly mutated - altering this bytecode - it leads to different operator strands and, consequently, different behaviors at runtime. Note, the bytecode contains no data strands; these are dynamically generated during execution by the operator strands.
The virtual machine includes dedicated instructions for IO that, when invoked from an operator strand, allow programs to interact with the outside world. Specifically, there is an instruction that pulls input data in, and another instruction that pushes data out. In this machine learning example, the aim is to develop an image classifier, so these instructions are used to feed raw pixel data into the virtual machine and then output the predicted label.
See Figure 3 for an illustrated example of how a program might be run to classify MNIST images.
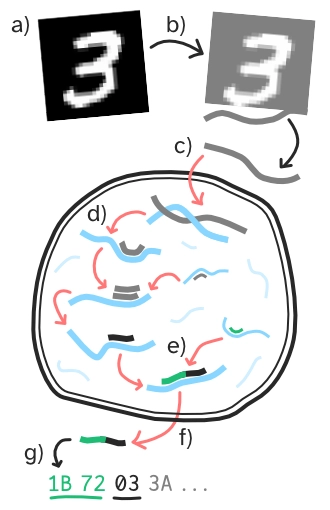
All the operator strands (depicted in blue in Figure 3), are executed concurrently, with the instruction encoded by each byte being evaluated sequentially. The instructions within the operator strands can perform a variety of actions, such as manipulating data strands, controlling program flow, or binding to other strands.
As the program runs, the operator strands process the initial input data, transforming it into a strand that corresponds to the output label (Figure 3d). To prevent short-circuiting (see initial program Methods 3.3), a two-byte magic number is added to the label-containing strand (Figure 3e, shown as the green strand). This modified strand is then output by the program (Figure 3f). The output is interpreted as a label (Figure 3g) by calculating the modulus 4 of the third byte. For example, an output value of 7 (7 % 4 = 3) would be interpreted as predicting the image as the digit 3.
3.2 Building the dataset
To simplify the task and accommodate the computational demands of the Zyme prototype implementation, we opted to use a tiny subset of the MNIST dataset. Our subset included only four classes (digits 0–3), with 520 training examples and 80 test examples, with each set containing an equal number of images of each digit. The raw images undergo preprocessing (as shown in Figure 3b) to accommodate a unique characteristic of the Zyme virtual machine: its distinctive byte architecture.
3.3 The initial program
The initial program (source) is hand-crafted and designed to operate as a crude hash function that reduces the pixel data of the input image into a single value, which will be interpreted as the predicted label. It employs a nested loop structure to iterate over every other row of the input image, processing each byte within those rows and adding them to a cumulative value. This accumulated value ultimately becomes the predicted label.
Before the final value is output, a magic number is pre-appended (as depicted in Figure 3). From previous small-scale experiments, we observed that initial programs can be easily ‘short-circuited’ by mutations causing them to prematurely return random values without processing the image data. While there is nothing inherently wrong with such programs, they often stagnate at random-level performance because they never process the input data, leaving no opportunity to learn from it. We enforce a minimal level of program complexity in the programs, by only considering candidate programs that include the correct magic number before the predicted label. Other programs are considered ‘dead’ and excluded during the selection stage.
While the resulting initial program is rudimentary - performing no better than random guessing, with an accuracy of 25% - this initial program consumes all of the input image and provides some material for evolution to build on. This program is written in Zyme source code, then compiled into bytecode consisting of 48 strands and a total size of 2.4kb.
3.4 Evolution and reproduction
Starting with a population consisting of 50 identical copies of the initial program, evolution follows a two step cycle: (1) reproduction with random mutation followed by (2) selection of the top-performing programs to form the next generation's population. In the first step (1), programs undergo reproduction and mutation. Reproduction occurs sexually, meaning two parent programs from the current population are randomly combined with crossover to create a new offspring program. Multiple mutations are then applied to this offspring, where random bytes within the bytecode are modified. In each generation, 500 pairs of programs are chosen (with replacement) from the population with a probability proportional to their accuracy in classifying images. Each pair produces 30 offspring programs, resulting in a total of 15,000 offspring programs per generation.
During the next step (2), each offspring program is evaluated using a batch of training examples, created at the start of each generation by randomly sampling 10 examples of each digit from the full training dataset. Each offspring program is run on every example in the batch. The offspring program's accuracy is calculated as the number of correctly predicted labels divided by the total number of trials (in this case, 40). From the pool of candidates, 50 programs are randomly chosen with a probability proportional to their accuracy. These 50 programs become the next generation's population.
4. Discussion
Genetic programming has long faced a paradoxical question: why can natural evolution produce such remarkable biodiversity while we struggle to evolve computer programs that solve even trivial tasks? Our work suggests the missing element may be specialised computer architectures optimised for evolution. Taking inspiration from biology, we developed Zyme, an evolution-oriented language and unique virtual machine. By evolving Zyme bytecode, we consistently discovered performance-enhancing mutations, observing population-wide accuracy improvements of ~25% over 50 generations, with exceptional individuals achieving ~40% gains on subsets of MNIST dataset.
While this study focused on image classification, our method reveals a path toward more generalizable genetic programming Zyme’s virtual machine's instruction set is general-purpose, facilitating only byte manipulation and control flow, with no specialized functions for image processing, suggesting the method may generalize to diverse machine learning tasks beyond computer vision. However, further research is clearly necessary. While we observe performance improvements, the underlying mechanisms are not yet fully understood; it is unclear whether these gains stem from meaningful algorithmic discoveries or merely minor optimizations of the initial hand-crafted program. That said, the evolved programs should be inherently interpretable, as they can be decompiled and their control flow directly analyzed (this capability has not yet been implemented).
Despite these open questions, our results make us optimistic that evolving Zyme bytecode could competitively solve the full MNIST task in the future.
1
Initial attempts at evolving programs were isolated from one another. Alan Turing, in his works Intelligent Machinery and Computing Machinery and Intelligence, envisioned automatic programming - a method by which computers could write programs through a trial-and-error process. However, he did little to realize this idea in practice. Around the same time, John von Neumann, as part of his work on cellular automata was exploring self-reproducing programs, and speculated whether if he mutated such programs they might evolve.
2
Taken from Genetic programming as a means for programming computers by natural Selection by John Koza in 1994.
3
While genetic programming has been applied to MNIST classification in prior work, existing approaches have yet to demonstrate a complete end-to-end solution. Several studies, such as Ruberto et al. in Image feature learning with genetic programming (2020) have employed genetic programming solely for feature extraction, while others like Torabi et al. in Using Cartesian Genetic Programming Approach with New Crossover Technique to Design Convolutional Neural Networks (2023) have used genetic programming to optimize neural network architectures. While these hybrid strategies have good performance for image classification, they lack scalability and generalizability, and do little to demonstrate the potential of genetic programming as a versatile, general-purpose machine learning method. A truly autonomous genetic programming system (as we hope to show here) should demonstrate the ability to: learn directly from raw input data without predefined feature extraction, and evolve complete, self-contained programs that process images and output predictions without relying on external machine learning components.
4
Although optimising for evolution have been explored in prior languages and virtual machines, their designs typically stem from traditional, human-oriented architectures, and thus, often encounter the aforementioned limitations. Notable examples include Lee Spector's Push programming language, which extends a stack-based machine by incorporating multiple separate stacks for each data type, allowing flexibility in manipulating functions with diverse type signatures.
5
Most existing genetic programming approaches to image classification incorporate substantial domain-specific knowledge, typically through either preprocessed image features or specialized operators designed for image manipulation rather than general byte-level operations. A representative example is Evans et al. in Evolutionary Deep Learning: A Genetic Programming Approach to Image Classification (2018) where the system directly implements convolutional and pooling operations as built-in functions but lacks support for dynamic or branching control flow. While this yields practical results, it constrains evolution to rearranging predefined functions rather than enabling the autonomous discovery of these high-level operations directly from raw data.
6
The concept of strand-based computational models - where strands are sequences of machine instructions that operate on one another - is not itself novel. Douglas Hofstadter introduced a mathematical puzzle called Typogenetics in Gödel, Escher, Bach (1979) to explore self-replication, and more recently, Stringmol demonstrated an artificial chemistry based on similar principles. However, this computational paradigm has never before been used as the foundation for a virtual machine.